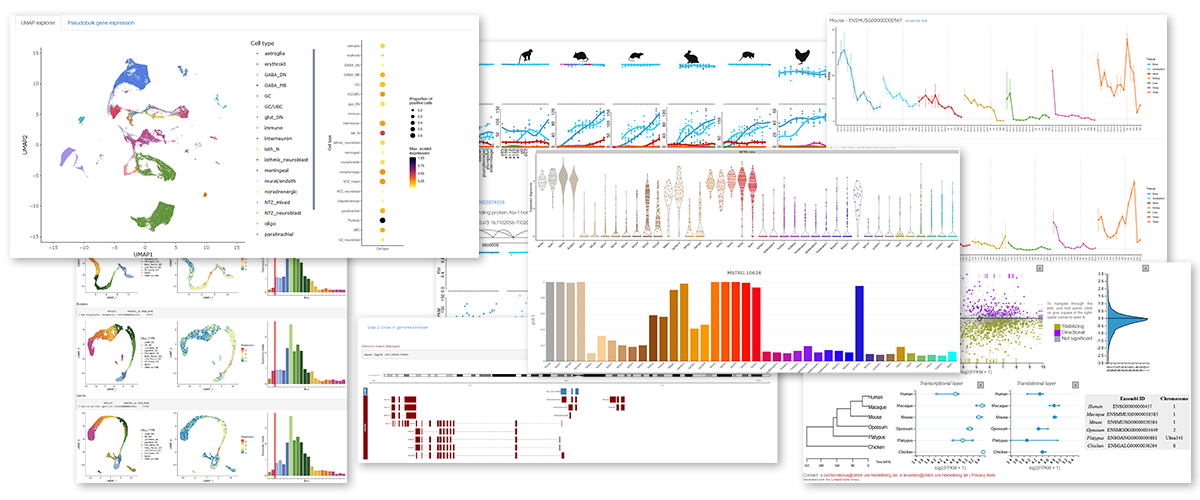
This Shiny app allows the interactive exploration of individual gene expression profiles across organs, developmental stages and species.
From the publication:
Cardoso-Moreira, M. et al. Gene expression across mammalian organ development. Nature (2019)
https://www.nature.com/articles/s41586-019-1338-5
This Shiny app allows the interactive exploration of lncRNA summary statistics, gene models and expression profiles across mammalian organs, developmental stages and species.
From the publication:
Sarropoulos, I. et al. Developmental dynamics of lncRNAs across mammalian organs and species. Nature (2019)
https://www.nature.com/articles/s41586-019-1341-x
This Shiny app allows the interactive exploration of alternative splicing patterns across mammalian organs, developmental stages and species.
From the publication:
Mazin, P.V. et al. Alternative splicing during mammalian organ development. Nat. Genet. (2021)
https://www.nature.com/articles/s41588-021-00851-w
From the publication:
Wang, Z.Y., Leushkin, E. et al. Transcriptome and translatome co-evolution in mammals. Nature (2020)
https://www.nature.com/articles/s41586-020-2899-z
This Shiny app allows the interactive exploration of individual gene expression profiles of adult testis across ten mammalian species and a bird at the single-cell resolution.
From the publication:
Murat F., Mbengue N. et al. The molecular evolution of spermatogenesis across mammals (2021)
https://www.nature.com/articles/s41586-022-05547-7
A comprehensive single-cell atlas of the sea-lamprey (Petromyzon marinus) brain.
From the publication:
Lamanna, F., Hervas-Sotomayor, F. et al. Reconstructing the ancestral vertebrate brain using a lamprey neural cell type atlas (2022)
https://www.biorxiv.org/content/10.1101/2022.02.28.482278v1
Explore the single nucleus RNA sequencing atlases of the human, mouse and opossum cerebellum, by Kaessmann Lab and Pfister Lab ( Datacommons ).
From the publication:
Sepp, M., Kevin, L. et al. Cellular development and evolution of the mammalian cerebellum. Nature (2023)
https://www.nature.com/articles/s41586-023-06884-x
This app allows the exploration of cis-regulatory elements (CREs) active in the developing mouse cerebellum.
From the publication:
Sarropoulos, I., Sepp M. et al. Developmental and evolutionary dynamics of cis-regulatory elements in mouse cerebellar cells. Science (2021)
https://www.science.org/doi/10.1126/science.abg4696
This Shiny app allows the interactive exploration of sex-biased gene expression profiles across organs, developmental stages and species.
From the publication:
Leticia Rodríguez-Montes et al. Sex-biased gene expression across mammalian organ development and evolution. Science (2023).
https://www.science.org/doi/10.1126/science.adf1046